Build a Decision Tree by Hand with TensorFlow Decision Forest

1 Problem Statement
In the real business world, business strategies are mostly generated by decision trees, but the native rules of a decision tree have some issues:
- Some features might lead to legal/compliance controversy or customer complaints
- The threshold of split features lack readability, such as 0.1483329203
In order to address these 2 issues, data scientists need to manually tweak nodes in a decision tree in order to meet business requirements, normally this process is done with commercial softwares as they provide flexibility for tree fine-tuning. But now, this can be done with TensorFlow Decision Forest in Python!
In this post, a way to build decision tree by hand with TensorFlow Decision Forest is shared so that data scientists can fusion the expertise with the native machine learning model to address business needs.
This is the summary of each step in this workflow:
- Build a native machine model
- Fine tune nodes to build a fusion tree
- Reset the performance of each leaf in the fusion tree
- Build a new model shell containing the fusion tree
- Use the new model to predict on the dataset and get prediction results
- Refresh the fusion tree with the literal performance of each leaf node
A public dataset for penguins species prediction is used in this post and the code has been tested in Google Colab as of July 2023.
2 Workflow
2.1 Prepare Dataset
Firstly, let's import some packages, note that the version of tensorflow_decision_forests is 1.5.0, make sure your TensorFlow is also compatible.
import tensorflow_decision_forests as tfdf
import os
import numpy as np
import pandas as pd
import tensorflow as tf
import matplotlib.pyplot as plt
import math
import collections
Then let's download the dataset by using the wget command and read it through Pandas:
# Download the dataset
!wget -q https://storage.googleapis.com/download.tensorflow.org/data/palmer_penguins/penguins.csv -O /tmp/penguins.csv
# Load a dataset into a Pandas Dataframe.
dataset_df = pd.read_csv("/tmp/penguins.csv")
The data is like this:
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | year | |
|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | male | 2007 |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | female | 2007 |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | female | 2007 |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN | 2007 |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | female | 2007 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | Chinstrap | Dream | 55.8 | 19.8 | 207.0 | 4000.0 | male | 2009 |
| 340 | Chinstrap | Dream | 43.5 | 18.1 | 202.0 | 3400.0 | female | 2009 |
| 341 | Chinstrap | Dream | 49.6 | 18.2 | 193.0 | 3775.0 | male | 2009 |
| 342 | Chinstrap | Dream | 50.8 | 19.0 | 210.0 | 4100.0 | male | 2009 |
| 343 | Chinstrap | Dream | 50.2 | 18.7 | 198.0 | 3775.0 | female | 2009 |
Then convert this into a TensorFlow Dataset:
# Convert the pandas dataframe into a tf dataset.
dataset_tf = tfdf.keras.pd_dataframe_to_tf_dataset(dataset_df, label="species")
2.2 Train a Decision Tree
# Train the decicion tree
model = tfdf.keras.CartModel()
model.fit(x=dataset_tf)
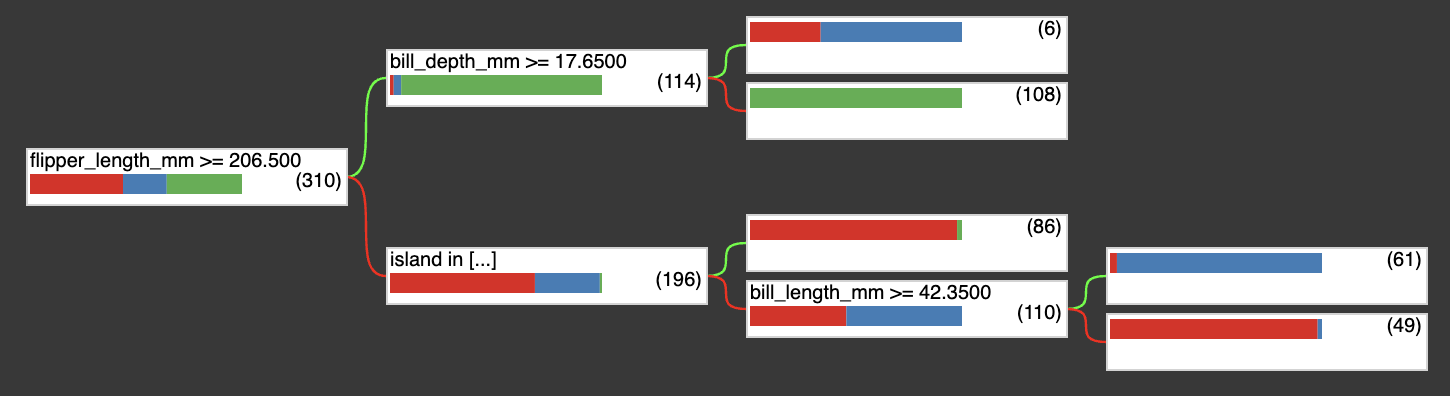
After training, you can plot the model this way:
tfdf.model_plotter.plot_model_in_colab(model, tree_idx=0, max_depth=5)
2.3 Fine Tune Nodes
This step shows how you can tweak some nodes of the tree above, fusion your expertise and customize your model. Firstly let's define some aliases that will be used later.
# Create some aliases
Tree = tfdf.py_tree.tree.Tree
SimpleColumnSpec = tfdf.py_tree.dataspec.SimpleColumnSpec
ColumnType = tfdf.py_tree.dataspec.ColumnType
# Nodes
NonLeafNode = tfdf.py_tree.node.NonLeafNode
LeafNode = tfdf.py_tree.node.LeafNode
# Conditions
NumericalHigherThanCondition = tfdf.py_tree.condition.NumericalHigherThanCondition
CategoricalIsInCondition = tfdf.py_tree.condition.CategoricalIsInCondition
# Leaf values
ProbabilityValue = tfdf.py_tree.value.ProbabilityValue
sample_tree = inspector.extract_tree(tree_idx=0)
print(sample_tree)
The output is like below, this is a plain text version of the tree plot above.
(flipper_length_mm >= 206.5; miss=False, score=0.5436033606529236)
├─(pos)─ (bill_depth_mm >= 17.649999618530273; miss=False, score=0.2061920464038849)
│ ├─(pos)─ ProbabilityValue([0.3333333333333333, 0.6666666666666666, 0.0],n=6.0) (idx=4)
│ └─(neg)─ ProbabilityValue([0.0, 0.0, 1.0],n=108.0) (idx=3)
└─(neg)─ (island in ['Biscoe', 'Torgersen']; miss=True, score=0.23399487137794495)
├─(pos)─ ProbabilityValue([0.9767441860465116, 0.0, 0.023255813953488372],n=86.0) (idx=2)
└─(neg)─ (bill_length_mm >= 42.349998474121094; miss=True, score=0.5646106004714966)
├─(pos)─ ProbabilityValue([0.03278688524590164, 0.9672131147540983, 0.0],n=61.0) (idx=1)
└─(neg)─ ProbabilityValue([0.9795918367346939, 0.02040816326530612, 0.0],n=49.0) (idx=0)
Then a node threshold of feature bill_depth_mm is updated to a value with better readability:
sample_tree.root.pos_child.condition = NumericalHigherThanCondition(
feature=SimpleColumnSpec(name="bill_depth_mm", type=ColumnType.NUMERICAL),
threshold=17.5,
missing_evaluation=False)
2.4 Tree Traversal
Traversal allows us to save each nodes of the tree, which is helpful for updating the tree in later steps, let's define a few traversal functions.
def leaf_reset_util(root, num_classes, scale_div):
"""
Reset the probability values of each leaf node.
Parameters
-----------
root: root node of a tree.
num_classes: number of classes.
scale_div: a numeric scale denominator used to create mutual exclusive probablity of each leaf node.
Return
-----------
A tree structure with reset probability for each leaf node.
"""
import tensorflow_decision_forests as tfdf
# Alias
LeafNode = tfdf.py_tree.node.LeafNode
ProbabilityValue = tfdf.py_tree.value.ProbabilityValue
if num_classes < 2:
raise ValueError("The number of unique classes should be at least 2 i.e."
" binary classification.")
if not isinstance(num_classes, int):
raise ValueError(f"The number of unique classes must be a integer greater than or equal to 2, got {num_classes}.")
if root is None:
return
if isinstance(root, LeafNode):
# make a pseudo probablity list based on number of classes
prob_list = [0 for i in range(num_classes)]
# assign the probablity of the last class as the leaf index divided by the scale denominator
prob_list[-1] = root.leaf_idx/scale_div
root.value = ProbabilityValue(probability=prob_list)
else:
leaf_reset_util(root.pos_child, num_classes, scale_div)
leaf_reset_util(root.neg_child, num_classes, scale_div)
return
def inoder_traverse(root, mode):
"""
Traverse the tree using in-order traversal.
Parameters
-----------
root: root node of a tree.
mode: the type of node to traverse.
Return
-----------
A list of nodes in the tree base on the choice of traversal mode.
"""
answer = []
inorder_traversal_util(root, answer, mode)
return answer
def inorder_traversal_util(root, answer, mode):
"""
Traverse the tree using in-order traversal.
Parameters
-----------
root: root node of a tree.
answer: a list placeholder to store the traversal answer.
mode: the type of node to traverse.
Return
-----------
A list of nodes in the tree base on the choice of traversal mode.
"""
if mode == 'all':
if root is None:
return
# if this is a leaf node, just return the value, otherwise, continue the traversal
if isinstance(root, LeafNode):
answer.append(root.value)
else:
inorder_traversal_util(root.pos_child, answer, mode)
answer.append(root.value)
inorder_traversal_util(root.neg_child, answer, mode)
return
elif mode == 'leaf':
if root is None:
return
# if this is a leaf node, just return the value, otherwise, continue the traversal
if isinstance(root, LeafNode):
answer.append([root.value.probability, root.value.num_examples, root.leaf_idx])
else:
inorder_traversal_util(root.pos_child, answer, mode)
inorder_traversal_util(root.neg_child, answer, mode)
return
else:
raise ValueError(f"Use either 'all' or 'leaf' mode to traverse, but got '{mode}', nothing is returned.")
Remember that the node of the tree has been modified while the prediction value of each node has not been updated. Thus, let's firstly reset the prediction value of each node, as a placeholder. This placeholder will be updated with literal performance of each node later.
scale_div = 100
leaf_reset_util(root=sample_tree.root,
num_classes=3,
scale_div=scale_div)
2.5 Tree Builder
In this step, the tree builder in TensorFlow Decision Forest package has been used to insert the updated tree into a real model, so that the model can be used for predicting and updating the placeholders.
Firstly, the orignal model's signature has been copied, and will be used in the builder to make sure that the new model handles the data types correctly, more context of this purpose can be found in the conversation with the TensorFlow Decision Forest author.
def copy_model_sig(model):
"""
Copy the model signature to a new model.
Parameters
-----------
model: a native model built by tfdf.keras.
Return
-----------
A copy of the original model's signature.
"""
spec = model.save_spec()[0][0]
return lambda insp: spec
classes = ["Adelie", "Gentoo" , "Chinstrap"]
classes.sort()
# Create the model builder
model_trial_idx = 0
model_trial_idx += 1
model_path = f"/tmp/manual_model/{model_trial_idx}"
!rm -rf /tmp/manual_model
builder = tfdf.builder.CARTBuilder(
path=model_path,
objective=tfdf.py_tree.objective.ClassificationObjective(
label="species", classes=classes),
input_signature_example_fn=copy_model_sig(model)
)
Then add the tree and conclude the building process:
builder.add_tree(sample_tree)
builder.close()
2.6 Activate the Fusion Model
Like stated earlier, the builder is able to create a regular Keras model for prediction, so let's active this.
fusion_model = tf.keras.models.load_model(model_path)
pred_value = fusion_model.predict(dataset_tf)
pred_value[:5]
The output is like:
array([[0. , 0. , 0.02],
[0. , 0. , 0.02],
[0. , 0. , 0.02],
[0. , 0. , 0.02],
[0. , 0. , 0.02]], dtype=float32)
The leaf assignment can be calculated by multiplying the prediction values with the scale divisor.
dataset_df['leaf_idx'] = pred_value[:,2]*scale_div
dataset_df
The output is like:
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | year | leaf_idx | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | male | 2007 | 2.0 |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | female | 2007 | 2.0 |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | female | 2007 | 2.0 |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN | 2007 | 2.0 |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | female | 2007 | 2.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | Chinstrap | Dream | 55.8 | 19.8 | 207.0 | 4000.0 | male | 2009 | 4.0 |
| 340 | Chinstrap | Dream | 43.5 | 18.1 | 202.0 | 3400.0 | female | 2009 | 1.0 |
| 341 | Chinstrap | Dream | 49.6 | 18.2 | 193.0 | 3775.0 | male | 2009 | 1.0 |
| 342 | Chinstrap | Dream | 50.8 | 19.0 | 210.0 | 4100.0 | male | 2009 | 4.0 |
| 343 | Chinstrap | Dream | 50.2 | 18.7 | 198.0 | 3775.0 | female | 2009 | 1.0 |
Next, let's calculate the probability and number of samples of each leaf.
leaf_num_samples = dataset_df.groupby('leaf_idx').agg(total_samples=('species','count')).reset_index()
node_prob_res = dataset_df.groupby(['leaf_idx','species']).agg(num_samples=('species','count')).unstack().stack(dropna=False).reset_index().fillna(0)
node_prob_res = node_prob_res.merge(leaf_num_samples, on='leaf_idx', how='left')
node_prob_res['prob'] = node_prob_res['num_samples']/node_prob_res['total_samples']
node_prob_array = node_prob_res.groupby('leaf_idx')['prob'].apply(list)
node_prob_res
| leaf_idx | species | num_samples | total_samples | prob | |
|---|---|---|---|---|---|
| 0 | 0.0 | Adelie | 53.0 | 54 | 0.981481 |
| 1 | 0.0 | Chinstrap | 1.0 | 54 | 0.018519 |
| 2 | 0.0 | Gentoo | 0.0 | 54 | 0.000000 |
| 3 | 1.0 | Adelie | 2.0 | 64 | 0.031250 |
| 4 | 1.0 | Chinstrap | 62.0 | 64 | 0.968750 |
| 5 | 1.0 | Gentoo | 0.0 | 64 | 0.000000 |
| 6 | 2.0 | Adelie | 95.0 | 97 | 0.979381 |
| 7 | 2.0 | Chinstrap | 0.0 | 97 | 0.000000 |
| 8 | 2.0 | Gentoo | 2.0 | 97 | 0.020619 |
| 9 | 3.0 | Adelie | 0.0 | 122 | 0.000000 |
| 10 | 3.0 | Chinstrap | 0.0 | 122 | 0.000000 |
| 11 | 3.0 | Gentoo | 122.0 | 122 | 1.000000 |
| 12 | 4.0 | Adelie | 2.0 | 7 | 0.285714 |
| 13 | 4.0 | Chinstrap | 5.0 | 7 | 0.714286 |
| 14 | 4.0 | Gentoo | 0.0 | 7 | 0.000000 |
2.7 Refresh the Fusion Model
Now that the literal performance of each nodes has been calculated, these results can be used to update the fusion model.
def leaf_res_refresh(root, probs, num_samples):
"""
Reset the probability values of each leaf node.
Parameters
-----------
root: root node of a tree.
probs: an array of probability of each class, for each node
num_samples: number of example in the node.
Return
-----------
A tree structure with literal probability and number of samples for each leaf node.
"""
import tensorflow_decision_forests as tfdf
# Alias
LeafNode = tfdf.py_tree.node.LeafNode
ProbabilityValue = tfdf.py_tree.value.ProbabilityValue
if root is None:
return
if isinstance(root, LeafNode):
root.value = ProbabilityValue(probability=probs[root.leaf_idx], num_examples=num_samples[root.leaf_idx])
else:
leaf_res_refresh(root.pos_child, probs, num_samples)
leaf_res_refresh(root.neg_child, probs, num_samples)
return
leaf_res_refresh(root=sample_tree.root,
probs=node_prob_array,
num_samples=leaf_num_samples['total_samples'])
print(sample_tree)
Then the tree has been updated:
(flipper_length_mm >= 206.5; miss=False, score=0.5436033606529236)
├─(pos)─ (bill_depth_mm >= 17.5; miss=False, score=None)
│ ├─(pos)─ ProbabilityValue([0.2857142857142857, 0.7142857142857143, 0.0],n=7) (idx=4)
│ └─(neg)─ ProbabilityValue([0.0, 0.0, 1.0],n=122) (idx=3)
└─(neg)─ (island in ['Biscoe', 'Torgersen']; miss=True, score=0.23399487137794495)
├─(pos)─ ProbabilityValue([0.979381443298969, 0.0, 0.020618556701030927],n=97) (idx=2)
└─(neg)─ (bill_length_mm >= 42.349998474121094; miss=True, score=0.5646106004714966)
├─(pos)─ ProbabilityValue([0.03125, 0.96875, 0.0],n=64) (idx=1)
└─(neg)─ ProbabilityValue([0.9814814814814815, 0.018518518518518517, 0.0],n=54) (idx=0)
Let's visualize the fusion model:
new_model_trial_idx = 2
new_model_trial_idx += 1
new_model_path = f"/tmp/manual_model/{new_model_trial_idx}"
!rm -rf /tmp/manual_model
new_builder = tfdf.builder.CARTBuilder(
path=new_model_path,
objective=tfdf.py_tree.objective.ClassificationObjective(
label="species", classes=classes),
input_signature_example_fn=copy_model_sig(model)
)
new_builder.add_tree(sample_tree)
new_builder.close()
new_fusion_model = tf.keras.models.load_model(new_model_path)
tfdf.model_plotter.plot_model_in_colab(new_fusion_model)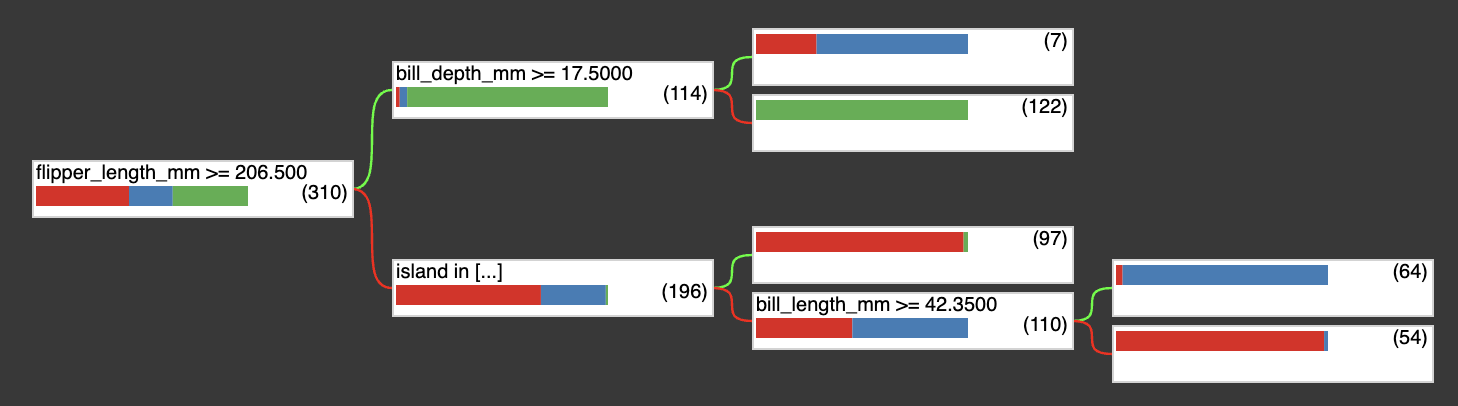
2.8 Extract the Tree Rules
Last step is to extract the rules of the tree so that these rules can be further fine-tuned by hand or used for deployment.
def get_tree_path(node):
"""
Parse a binary tree to get all decision paths.
Parameters:
-----------
node: root node of the tree structure to traverse.
Return:
-----------
All decision path from root to each leaf node.
"""
import tensorflow_decision_forests as tfdf
# Alias
LeafNode = tfdf.py_tree.node.LeafNode
CategoricalIsInCondition = tfdf.py_tree.condition.CategoricalIsInCondition
paths = []
path = []
def recurse_(node, path, paths):
if not isinstance(node, LeafNode):
# Categorical features
if isinstance(node.condition, CategoricalIsInCondition):
name = node.condition.feature.name
p1, p2 = list(path), list(path)
p1 += [f"({name} in {str(node.condition.mask)})"]
recurse_(node.neg_child, p1, paths)
p2 += [f"({name} not in {str(node.condition.mask)})"]
recurse_(node.pos_child, p2, paths)
# Numerical features
else:
name = node.condition.feature.name
threshold = float(node.condition.threshold)
p1, p2 = list(path), list(path)
p1 += [f"({name} < {threshold})"]
recurse_(node.neg_child, p1, paths)
p2 += [f"({name} >= {threshold})"]
recurse_(node.pos_child, p2, paths)
else:
path += [(node.value.probability, node.value.num_examples, node.leaf_idx)]
paths += [path]
recurse_(node=node,
path=path,
paths=paths)
rule_dict = {}
# Format the results
for path in paths:
rule_path = []
num_samples = path[-1][1]
leaf_idx = path[-1][-1]
for p in path[:-1]:
rule_path.append(str(p))
leaf_probability = path[-1][0]
rule_result = [rule_path, leaf_probability, num_samples]
rule_dict[leaf_idx] = rule_result
return rule_dictrule_res = get_tree_path(sample_tree.root)When this function is called, it returns a dictionary {leaf index: [rules, probability, number of samples]}.
{0: [['(flipper_length_mm < 206.5)',
"(island in ['Biscoe', 'Torgersen'])",
'(bill_length_mm < 42.349998474121094)'],
[0.9814814814814815, 0.018518518518518517, 0.0],
54],
1: [['(flipper_length_mm < 206.5)',
"(island in ['Biscoe', 'Torgersen'])",
'(bill_length_mm >= 42.349998474121094)'],
[0.03125, 0.96875, 0.0],
64],
2: [['(flipper_length_mm < 206.5)',
"(island not in ['Biscoe', 'Torgersen'])"],
[0.979381443298969, 0.0, 0.020618556701030927],
97],
3: [['(flipper_length_mm >= 206.5)', '(bill_depth_mm < 17.5)'],
[0.0, 0.0, 1.0],
122],
4: [['(flipper_length_mm >= 206.5)', '(bill_depth_mm >= 17.5)'],
[0.2857142857142857, 0.7142857142857143, 0.0],
7]}3 Conclusion
As you can see, by utilizing TensorFlow Decision Forest builder along with custom traversal functions, you are able to
- Fusion your human expertise with native decision tree.
- Get the performance of your fusion model.
- Extract the decision rules programatically.
This solution is helpful for many data scientists who need to build rule-based business strategies which are mostly required to intuitive, compliant with laws and regulations.
The complete Colab notebook can be found below:

Build Decision Tree by Hand
Hopefully it helps.
